2025年发表文章
1. Shu-Yu Yang#, Ti-Ti Ying#, Tian-Hui Zhou, Yu-Tian Guan, Xu-Liang Xu, Hong Wang*, Bin Wei*. The Myxobacterial Genus Archangium: A Prolific and Underexploited Source of Bioactive Secondary Metabolites. Journal of Medicinal Chemistry, 2025, 68(3): 2183–2197. (https://doi.org/10.1021/acs.jmedchem.4c02203) IF:6.8
Abstract

The genus Archangium, a cryptic group of myxobacteria, is a rich source of diverse secondary metabolites. This study reviews the chemical structures and discovery history of 55 secondary metabolites, analyzing the relationship between the chemical structures of these compounds and their bioactivity profiles through molecular networking. Notably, 63.6% of the compounds exhibit potent antimicrobial (MIC < 1 μg/mL) and/or cytotoxic activities (IC50 < 1 μg/mL). Advances in the biosynthetic gene clusters and biosynthetic pathways of seven classes of identified compounds are also presented. Finally, genomic mining approaches are applied to analyze the potential for Archangium strains to synthesize analogs of identified bioactive natural products, uncovering that 98.7% of their secondary metabolic potential remains unexplored. This study highlights the vast potential of Archangium bacteria in synthesizing clade-specific novel secondary metabolites, particularly ribosomally synthesized and post-translationally modified peptide natural products, offering valuable insights for the targeted discovery and biosynthesis of new natural products from this genus.
2. Bin Wei#,*, Ti-Ti Ying#, Hua-Wei Lv#, Zhen-Yi Zhou, Hai Cai, Gang-Ao Hu, Hui-Min Liang, Wen-Chao Yu, Yan-Lei Yu, Ai-Li Fan, Kui Hong, Xing-Nuo Li*, Hong Wang*. Global analysis of fungal biosynthetic gene clusters reveals the diversification of diketopiperazine biosynthesis. Bioresource Technology, 2025, 422: 132218. (https://doi.org/10.1016/j.biortech.2025.132218) IF:9.7
Abstract
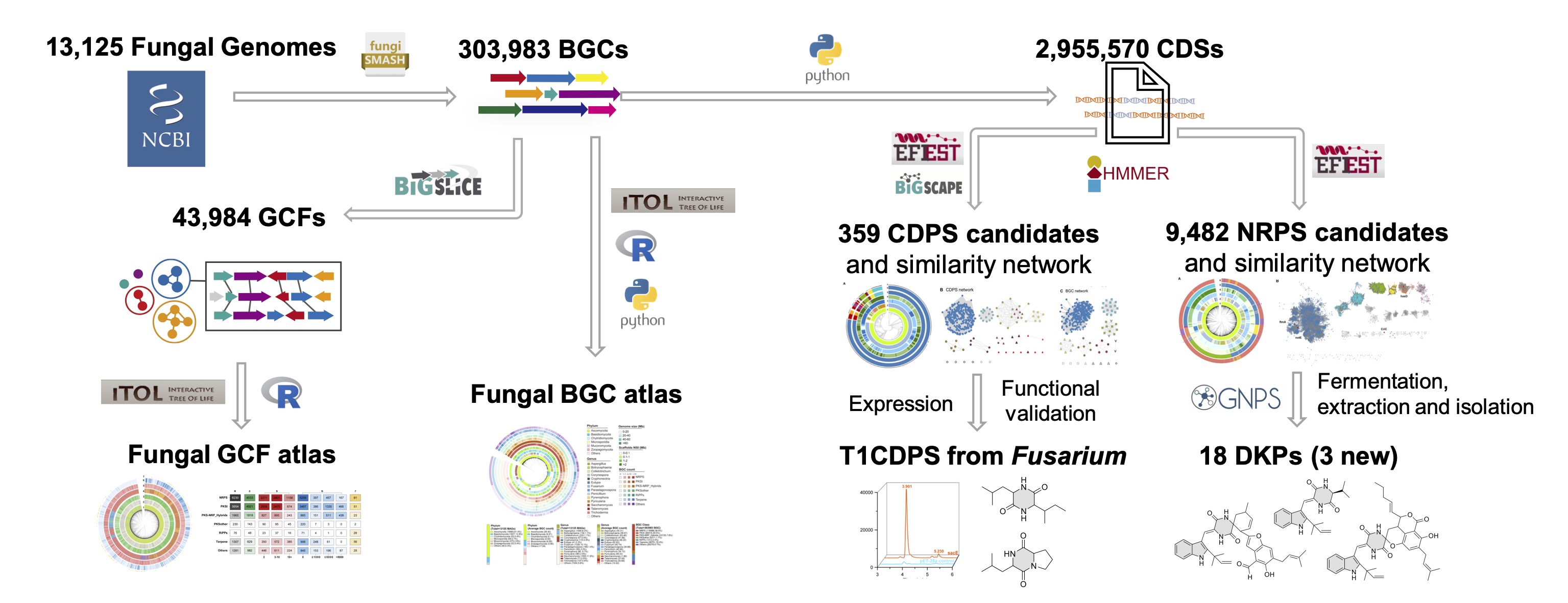
Fungi are a vast reservoir of structurally diverse natural products, yet their biosynthetic potential remains underexplored. Here, we present the most comprehensive fungal biosynthetic gene cluster (BGC) atlas, comprising 303,983 BGCs predicted from 13,125 fungal genomes, revealing numerous underexplored taxa harboring extensive biosynthetic diversity. These BGCs were classified into 43,984 gene cluster families (GCFs), of which 99.6 % remain uncharacterized. Gene-centric analysis revealed the presence of 359 cyclodipeptide synthases (CDPSs) of three distinct subcategories and 9,482 nonribosomal peptide synthetases (NRPSs) responsible for diketopiperazine biosynthesis in the fungal BGC atlas. Notably, 304 type I CDPSs were exclusively found in Fusarium, with one confirmed to be responsible for the synthesis of cyclo(Leu-Ile) and cyclo(Pro-Leu). Bioinformatics analysis suggests that three newly identified indole diketopiperazine alkaloids, isolated from a marine-derived Aspergillus strain, are synthesized by an NRPS. This study presents the most comprehensive fungal BGC atlas and highlights the diversification of diketopiperazine biosynthesis in fungi.
3. Quan Xu, Ye-Qing Du, Pan-Pan Chen, Yili Sun, Ze-Nan Yang, Hui Zhang, Bencan Tang, Hong Wang, Jia Li*, Yue-Wei Guo*, Xu-Wen Li*. Computation assisted chemical study of photo-induced late-stage skeleton transformation of marine natural products towards new scaffolds with biological functions. Chinese Chemical Letters, 2025, 36(5): 110141. (https://www.sciencedirect.com/science/article/pii/S1001841724006600?dgcid=coauthor) IF:9.4
Abstract

A computer-assisted chemical investigation of an intriguing photoreaction of norditerpenoids (3‒7) has been first reported, leading to not only their biomimetic conversion, but also the generation of several new products with uncommon 4,14-dioxabicyclo[10.2.1]pentadecane scaffold (8, 9, 12‒14). In bioassay, compounds 10 and 15 exhibited significant stimulation of GLP-1 secretion. This study has given an insight for the application of computational methods on the late-stage skeleton transformation of complex natural products towards new bioactive compounds.
4. Yanlei Yu#, Honggang Wang#, Xiaoshu Jin#, Wenjing Huang, Yunjie Zhao, Ningning Wang, Dongze Lu, Bin Wei, Hong Wang*. Structural characterization of Dendrobium officinale polysaccharides and their regulation effect on intestinal microbiota during in vitro fermentation. Polymers, 2025, 17(6): 727. (https://doi.org/10.3390/polym17060727) IF:4.7
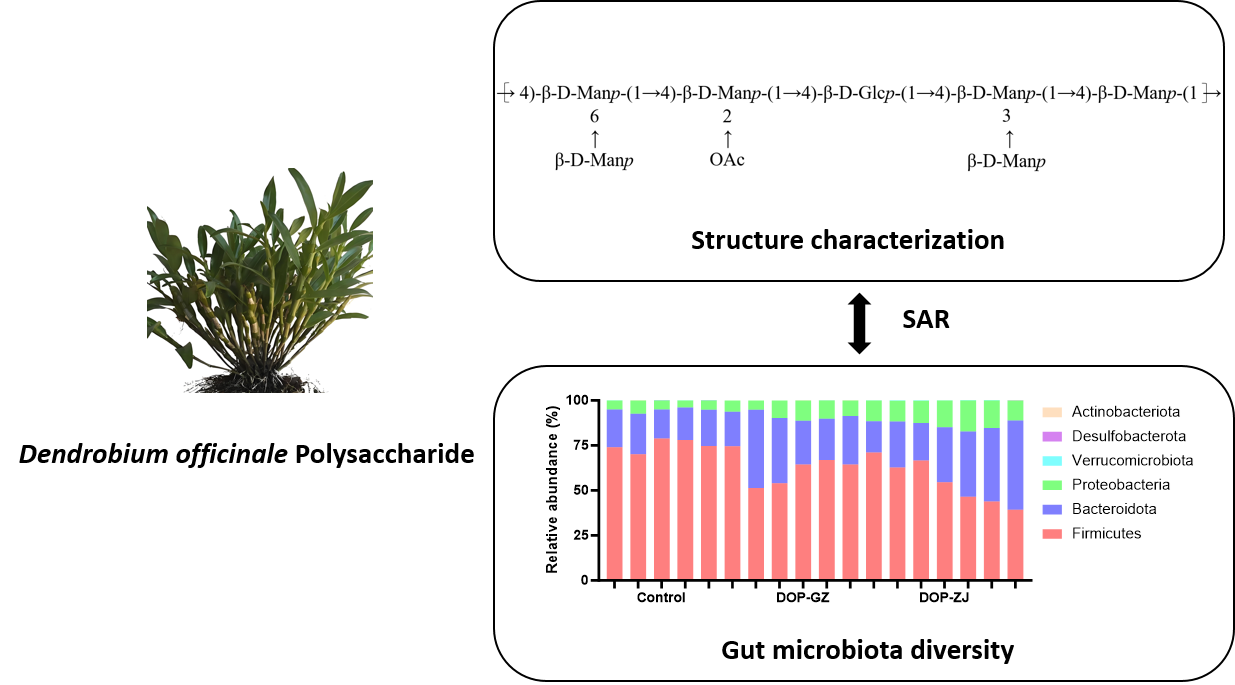
Polysaccharides derived from Dendrobium officinale have been demonstrated to exhibit metabolic regulatory properties. However, the correlation between their structure and function, particularly their mechanism of action through gut microbiota, remains underexplored. This study systematically elucidates the structural characteristics of Dendrobium officinale polysaccharide (DOP) from the Guizhou (GZ) and Zhejiang (ZJ) provinces of China using nuclear magnetic resonance (NMR) and a series of chromatographic analyses, revealing their unique molecular features. Additionally, the metabolic regulatory activities were assessed through α-glucosidase inhibitory assay and in vitro intestinal flora activity assay. The findings not only uncover a novel mechanism by which DOPs regulate metabolism through gut microbiota but also provide a crucial theoretical basis for the application of DOPs in functional foods and pharmaceutical development.
5. Buddha Bahadur Basnet, Zhen-Yi Zhou, Bin Wei*, Hong Wang*. Advances in AI-based strategies and tools to facilitate natural product and drug development. Critical Reviews in Biotechnology, 2025, 1-32. (https://doi.org/10.1080/07388551.2025.2478094) IF:8.2
Abstract
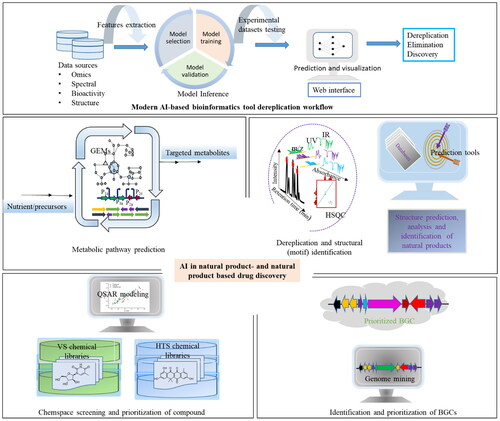
Natural products and their derivatives have been important for treating diseases in humans, animals, and plants. However, discovering new structures from natural sources is still challenging. In recent years, artificial intelligence (AI) has greatly aided the discovery and development of natural products and drugs. AI facilitates to: connect genetic data to chemical structures or vice-versa, repurpose known natural products, predict metabolic pathways, and design and optimize metabolites biosynthesis. More recently, the emergence and improvement in neural networks such as deep learning and ensemble automated web based bioinformatics platforms have sped up the discovery process. Meanwhile, AI also improves the identification and structure elucidation of unknown compounds from raw data like mass spectrometry and nuclear magnetic resonance. This article reviews these AI-driven methods and tools, highlighting their practical applications and guide for efficient natural product discovery and drug development.
6. Ti-Ti Ying#, Hao-Qiang Hu#, Xiao-Wen Wu, Xu-Liang Xu, Jian Lv, Shu-Ning Zhang, Hong Wang, Wei Hou*, Bin Wei*, Guo-Wu Rao*. Optimized ebselen derivatives as novel potent Escherichia coli β-glucuronidase covalent allosteric inhibitors. European Journal of Medicinal Chemistry, 2025, 290: 117571. (https://doi.org/10.1016/j.ejmech.2025.117571) IF: 6.0
Abstract
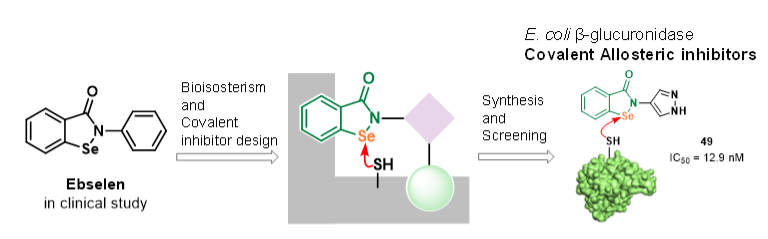
Gut microbial β-glucuronidase (GUS) plays a key role in metabolizing compounds and influencing disease and drug metabolism, highlighting the need for potent inhibitors to improve drug efficacy and intestinal health. To identify Escherichia coliβ-glucuronidase (EcGUS) inhibitors, we designed and synthesized fifty 1,2-benzoselenazol-3-one (BSEA) derivatives using a bioisosterism strategy. Among these, twenty-five BSEA derivatives demonstrated greater inhibitory efficacy than the most potent known EcGUS inhibitor, amoxapine (AMX), with compound 49showing the strongest activity, achieving an IC50 of 12.9 nM. Structure–inhibitory activity relationship analysis suggested that modifications such as adding benzene rings or nitrogenous heterocycles to the BSEA scaffold enhanced inhibitory activity, influenced by the type and position of substituents. The LC−MS analysis confirmed that compounds 31 and 49 covalently modify Cys197 in EcGUS, and additional covalent linkage of compound 49 was observed on Cys28 and Cys443. In addition, the jump dilution assays proved that compounds 31 was irreversible covalent inhibitors, and its kinetic parameter kinact/KI were determined to be 21292.9 M−1s−1. The compounds 49 was reversible covalent inhibitors and its apparent steady-state inhibition constant Ki∗app were determined to be 23.33 nM. Molecular docking predicted specific interactions, such as hydrogen bonds involving Se and the pyrazole NH of compound 49 with Cys28 and Cys449, which may contribute to its inhibitory action. This study reports the first discovery of covalent inhibitors for EcGUS, with optimized BSEA derivatives acting as novel allosteric covalent inhibitors, revealing structure-activity relationships and molecular determinants that establish their potential in drug development.C
7. Yi Hua,# Gangao Hu,# Titi Ying, Jiangwei Pan, Zhenyi Zhou, Cuixian Zhang,* Hong Wang,* and Bin Wei*. The Pathogenic Fungal Genus Corynespora: A Prolific Source of Bioactive Secondary Metabolites. Journal of Natural Products, 2025, 88 (5): 1259-1269. (https://doi.org/10.1021/acs.jnatprod.5c00145) IF:3.6
AbstractC

In this perspective, we examine the chemical structures and biological activities of 55 secondary metabolites isolated from the pathogenic fungal genus Corynespora. Over 20% of these compounds demonstrate bioactivities, including antibacterial, anticancer, enzyme inhibition, and others. Bioinformatic analysis reveals that Corynespora possesses a high potential for encoding PKS-I and NRPS natural products, including many clade-specific metabolites, and only 2.1% of biosynthetic gene cluster families are associated with known secondary metabolites. This study underscores the significant, unexplored potential of Corynespora strains for synthesizing novel secondary metabolites, providing valuable insights into the targeted discovery and biosynthesis of novel natural products from this genus.
8. Cai-Ling Yang#, Pan-Pan Wang#, Zhen-Yi Zhou, Xiao-Wen Wu, Yi Hua, Jian-Wei Chen, Hong Wang*, Bin Wei*. Discovery of naturally inspired antimicrobial peptides using deep learning. Bioorganic Chemistry, 2025, 160: 108444. (https://doi.org/10.1016/j.bioorg.2025.108444) IF: 4.7
Abstract
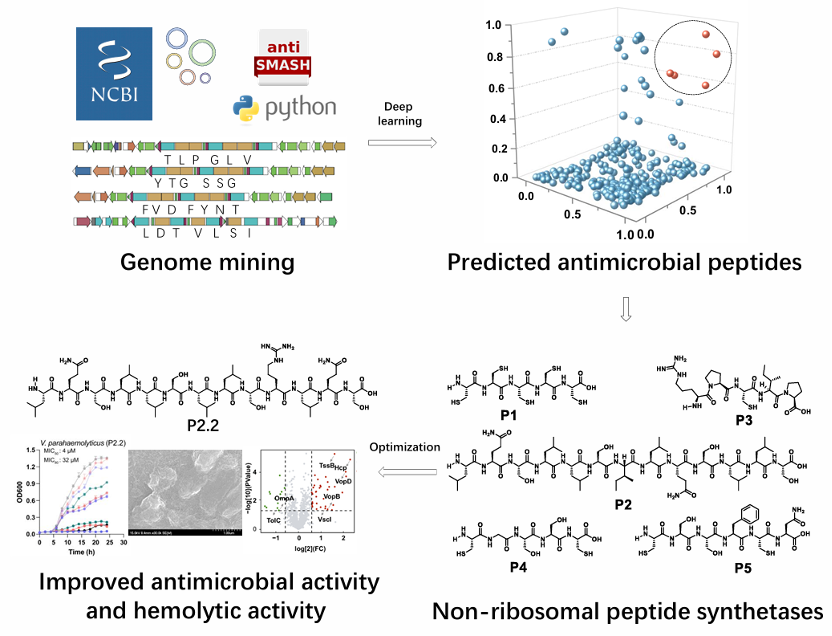
Non-ribosomal peptides (NRPs) are promising lead compounds for novel antibiotics. Bioinformatic mining of silent microbial NRPS gene clusters provide crucial insights for the discovery and de novo design of bioactive peptides. Here, we describe the efficient discovery and antibacterial evaluation of novel peptides inspired by metabolite scaffolds encoded by NRPS gene clusters from 216,408 bacterial genomes. In total, 335,024 NRPS gene clusters were identified and dereplicated, yielding 328 unique peptide scaffolds. Using deep learning-based scoring, five antimicrobial peptide candidates (P1–P5) were synthesized via solid-phase chemical synthesis. Among them, peptide P2 exhibited potent antibacterial activity with MIC50 values of 1–2 μM against two pathogenic strains. Subsequent amino acid optimization guided by deep learning algorithms produced P2.2, a derivative with significantly enhanced antibacterial activity. Mechanistic studies revealed that P2.2 disrupts bacterial membranes and increases permeability by modulating proteins involved in the type VI and III secretion systems. Furthermore, P2.2 demonstrated synergistic effects when combined with conventional antibiotics and exhibited reduced hemolytic activity, improving its therapeutic potential. These findings underscore the immense potential of deep learning to accelerate the discovery of naturally inspired antimicrobial peptides from silent biosynthetic gene clusters.
9. Bing-Cheng Dong#, Fen Liu#, Gang-Ao Hu, Wen-Chao Yu, Zi-Yang Li, Yu-Tian Guan, Wen-Wen Zhang, Hong Wang*, Bin Wei*. Metabologenomics analysis reveals antibiotic crypticity of Kutzneria viridogrisea DSM 43850. Journal of Applied Microbiology, 2025, 5(136): 114. (https://doi.org/10.1093/jambio/lxaf114) IF: 3.2
Abstract
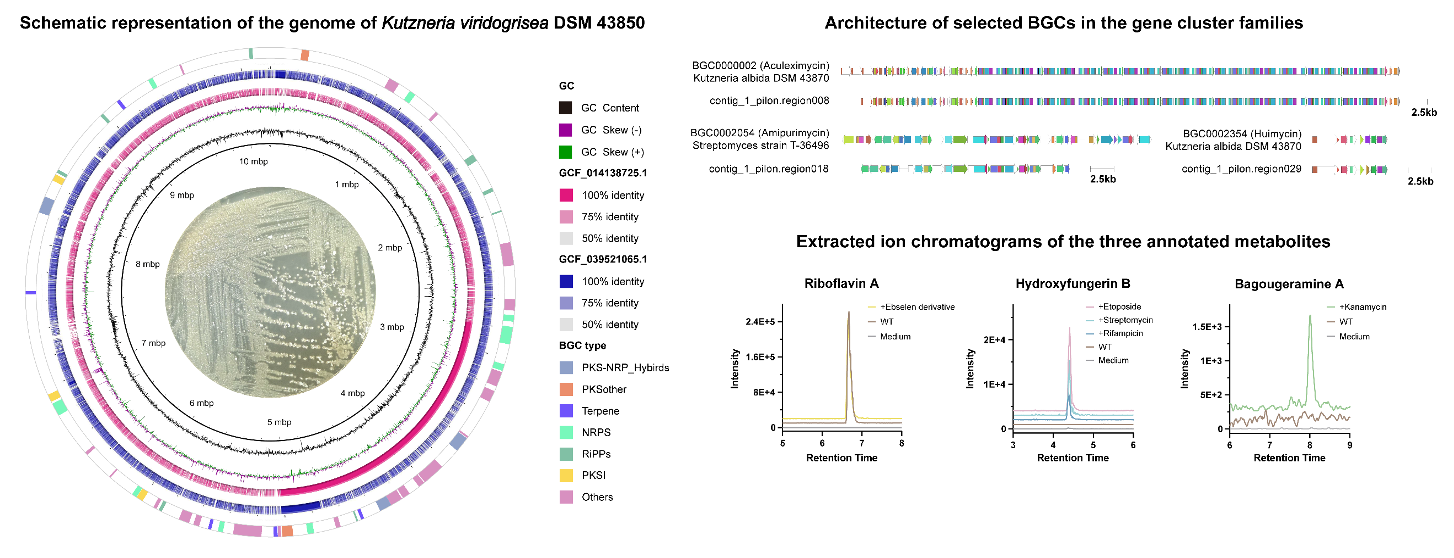
Aims This study aimed to explore the secondary metabolic potential of Kutzneria viridogrisea DSM 43850 by conducting whole-genome sequencing and utilizing bioinformatics tools to analyze its biosynthetic gene clusters (BGCs). Additionally, the secondary metabolites produced by this strain were investigated under various chemical elicitors using untargeted metabolomics techniques.
Methods and Results The complete genome of Kutzneria viridogrisea DSM 43850 was obtained by re-sequencing, followed by in-depth bioinformatics analysis to assess its secondary metabolic potential. The genome was found to encode a circular 10.2 Mb chromosome, with 4.3% of its functional genes involved in secondary metabolism. The strain harbors 52 BGCs, of which only four are associated with known products. Among these, eight gene clusters were identified as RiPPs (Ribosomally synthesized and post-translationally modified peptides), and the precursor peptide structures of four were predicted, all featuring novel scaffolds. Untargeted metabolomics analysis using LC-MS revealed that the strain could produce a series of novel secondary metabolites when induced with kanamycin and an ebselen derivative.
Conclusions This study highlights the significant secondary metabolic potential of Kutzneria viridogrisea DSM 43850, uncovering several novel BGCs and metabolic products.
10. Tongqing Li, Xueying Liu, Haifeng Qian, Sheyu Zhang, Yu Hou, Yuchao Zhang, Guoyan Luo, Xun Zhu, Yanxin Tao, Mengyang Fan, Hong Wang, Chulin Sha, Ailan Lin, Jingjing Qin, Kedan Gu, Weichang Chen, Ting Fu, Yajun Wang, Yong Wei, Qin Wu & Weihong Tan. Blocker-SELEX: a structure-guided strategy for developing inhibitory aptamers disrupting undruggable transcription factor interactions. Nature Communications, 2024, 15: 6751. ( https://www.nature.com/articles/s41467-024-51197-w) IF: 15.7
Abstract
Despite the well-established significance of transcription factors (TFs) in pathogenesis, their utilization as pharmacological targets has been limited by the inherent challenges in modulating their protein interactions. The lack of defined small-molecule binding pockets and the nuclear localization of TFs do not favor the use of traditional tools. Aptamers possess large molecular weights, expansive blocking surfaces and efficient cellular internalization, making them compelling tools for modulating TF interactions. Here, we report a structure-guided design strategy called Blocker-SELEX to develop inhibitory aptamers (iAptamers) that selectively block TF interactions. Our approach leads to the discovery of iAptamers that cooperatively disrupt SCAF4/SCAF8-RNAP2 interactions, dysregulating RNAP2-dependent gene expression, which impairs cell proliferation. This approach is further applied to develop iAptamers blocking WDR5-MYC interactions. Overall, our study highlights the potential of iAptamers in disrupting pathogenic TF interactions, implicating their potential utility in studying the biological functions of TF interactions and in nucleic acids drug discovery.
11. Zhehui Cai, Zhe Hu, Xiaojun Yao, Pengyu Liang, Huawei Zhang*. Single-crystal structure, Hirshfeld surface and DFT analyses of chrysomycin A, a marine microbe-derived promising agent for anticancer and antimicrobial therapy. Journal of Molecular Structure, 2025,1322: 140488. (https://doi.org/10.1016/j.molstruc.2024.140488) IF: 4.7
Abstract
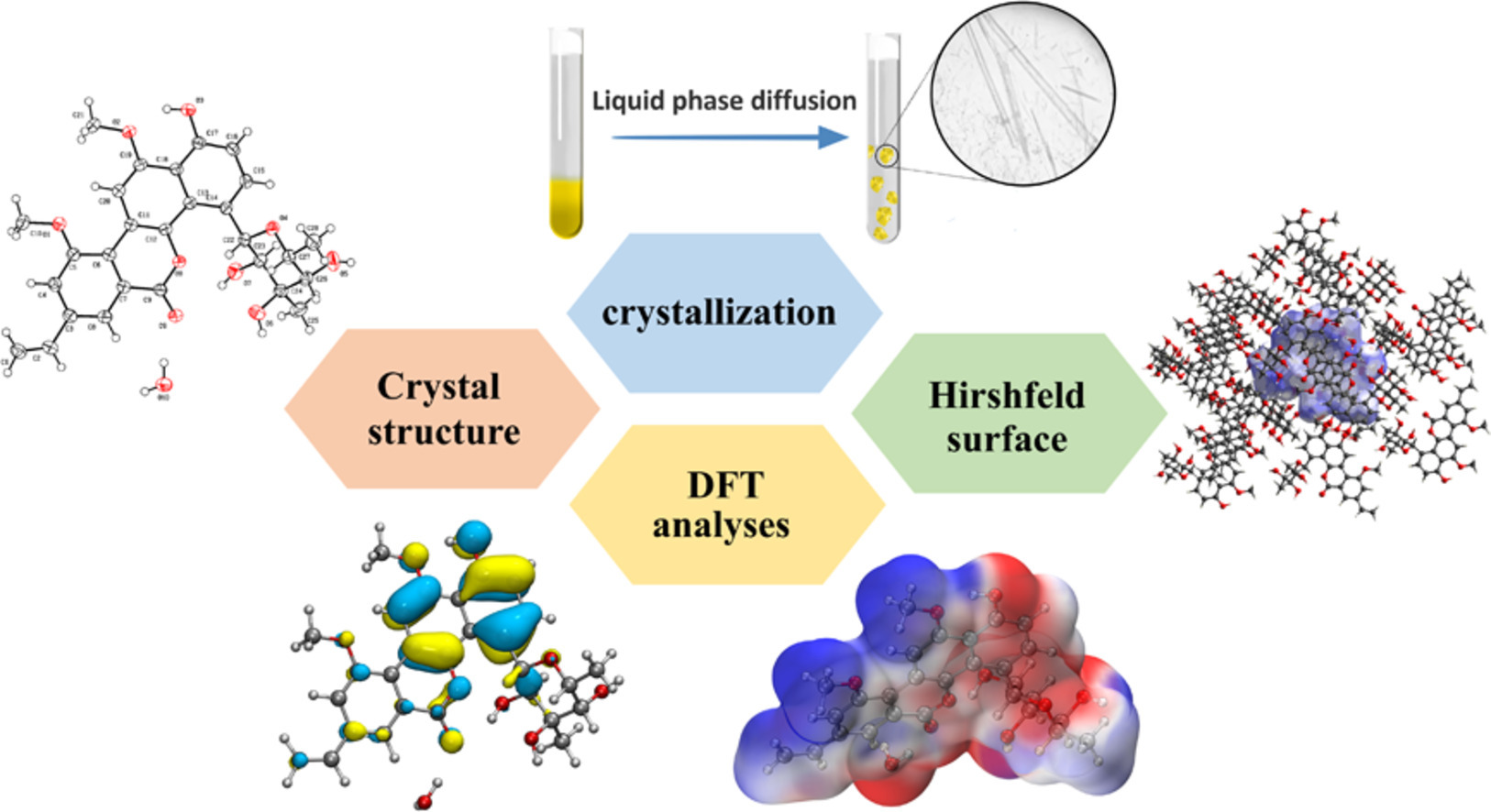
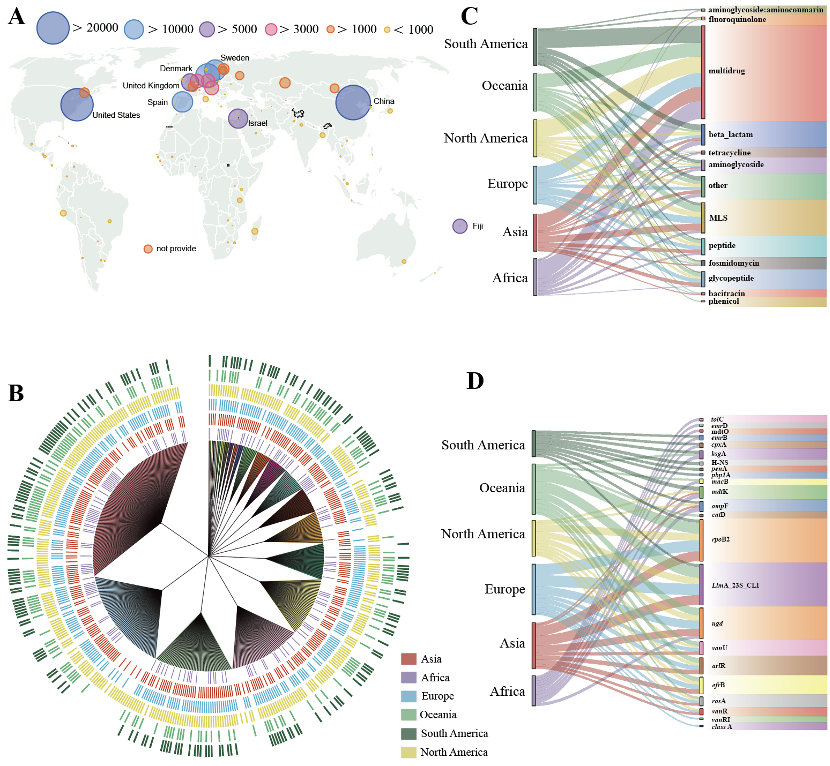
Chrysomycin A (CA) is an important Streptomyces-derived aromatic polyketide with therapeutic potential for treatment of cancer and bacterial infections. In order to prepare highly pure CA for new drug development, a simple and effective strategy to crystallize CA was firstly developed by the liquid phase diffusion method in this work. The single-crystal structure of CA and its Hirshfeld surface property analyses showed that the asymmetric unit of CA crystal consists of one CA molecule and a water in the orthorhombic space group P212121 and it has π-π interactions between benzene rings. CA crystal stacking is consolidated by three intermolecular hydrogen bonds including carbonyl/keto-C-O⋅⋅⋅H(water), naphthalene-C-O⋅⋅⋅H(water) and glycoside-O H⋅⋅⋅O(water) and van der Waals forces. Furthermore, the HOMO-LUMO energy gap, UV-vis spectral parameters, electrostatic potential, mullliken charge and nonlinear optical properties analyzed in DFT calculations. These results pave a fundamental way for the production of CA using the feasible crystallization approach and the study of molecular mechanisms of action (MoA).
12. Yifan Bai, Tao Luo, Zhehui Cai, Wangjie Zhu, Yuanyuan Liu, Huawei Zhang*. Green and efficient extraction of an anticaner agent N-methylsansalvamide using ultrasound-assisted deep eutectic solvents from mycelia of strain Frusarium sp. R1. Talanta, 2025, 282: 127017. (https://doi.org/10.1016/j.talanta.2024.127017) IF: 6.1
Abstract
N-methylsansalvamide (SA), one of cyclic pentadepsipeptides produced by several Fusarium strains, is a promising therapeutic agent for the treatment of cancer disease. In order to make sufficient amount of SA for drug development, a green and efficient extraction process of SA from the mycelia of strain Fusarium sp. R1 using deep eutectic solvent-assisted ultrasound extraction (DES-UAE) was firstly achieved in this work. Solvent screening results indicated that choline chloride-acetic acid (ChCl-Aa) was shown to be the best DES for SA extraction. Through single-factor trials, Plackett-Burman design (PBD) and BoxBehnken design (BBD) experiments, the optimal conditions for DES-UAE with the highest SA yield of 58.2 ± 1.1 mg/g were obtained as follows: ChCl-Aa ratio of 1:2.0 (M/M), water content of 16.4 %, liquid-solid ratio of 37:1 (mL/g), ultrasonic power of 175 W for 47.4 min at 46.3 °C. Compared to conventional extraction approaches, DES-UAE exhibited better SA yield since it caused more serious damage to the surface of mycelia powder on basis of scanning electron microscopy (SEM) analysis. Furthermore, molecular interaction studies suggested that SA has a variety of interactions with ChCl-Aa, including hydrogen and electrovalent bonds as well as van der Waals forces. Finally, the recovery rate of SA reached up to 99.5 % when the ratio of distilled water and DES extracts was 15:1 (V/V). These findings provide the way for large-scale production of SA.

13. Jiapei Chen, Xusheng Duan, Mengyuan Wu, Guishun Bai, Jianwei Chen, Xuanrong Sun, Jean Rodriguez, Damien Bonne*, Hong Wang* and Xiaoze Bao*. Enantioselective and Diastereodivergent Construction of Oxindole-pyrazolone Conjugates Bearing an Alkenyl Substituted Quaternary Chlorinated Stereogenic Centre. Organic Chemistry Frontiers, 2025. (https://doi.org/10.1039/D5QO00499C) IF: 4.7
Abstract
Chlorinated stereogenic carbon centres are important elements both in pharmaceutical reagents and synthetic intermediates. Herein, a novel methodology is reported for the construction of a rarely developed alkenyl substituted quaternary chlorinated stereogenic centre, featured in oxindole and pyrazolone pharmacophores. Remarkably, the configuration of the double bond was switchable via the combination of a suitable base and solvent. In addition, the enantioselective synthesis of Z-type products was achieved with natural quinidine as a catalyst, affording the chlorinated products in excellent yields and stereoselectivities. Preliminary 1H-NMR titration was studied to gain insights into the control of the double bond's configuration. Moreover, the anti-tumour activity against the A549 cell-line of these newly synthesized chemical entities was evaluated, and the product (E)-5ca was revealed to be a promising anti-tumour agent.
14. Yujie Zhao, Qing Gong, and Huawei Zhang*. Engineering of Global Transcriptional Regulators (GTRs) in Aspergillus for Natural Product Discovery. Journal of fungi, 2025, 11(6): 449. (https://doi.org/10.3390/jof11060449) IF: 4.0
Abstract
The Aspergillus genus is an important group of filamentous fungi, and the various biological activities of its secondary metabolites (SMs) have great biosynthetic potential. Despite over 4200 SMs having been isolated from Aspergillus spp., their metabolic potential remains unexplored due to the presence of numerous silent biosynthetic gene clusters (BGCs) in their genomes. Fortunately, over the last two decades, the global transcriptional regulator (GTR) engineering strategy has emerged as a powerful tool for activating these cryptic BGCs in order to synthesize previously undiscovered SMs from Aspergillus spp. This review highlights recent advances in fungal GTR engineering techniques, the regulatory mechanisms of GTRs, and current challenges and future perspectives for their application in natural product discovery in the genus Aspergillus.
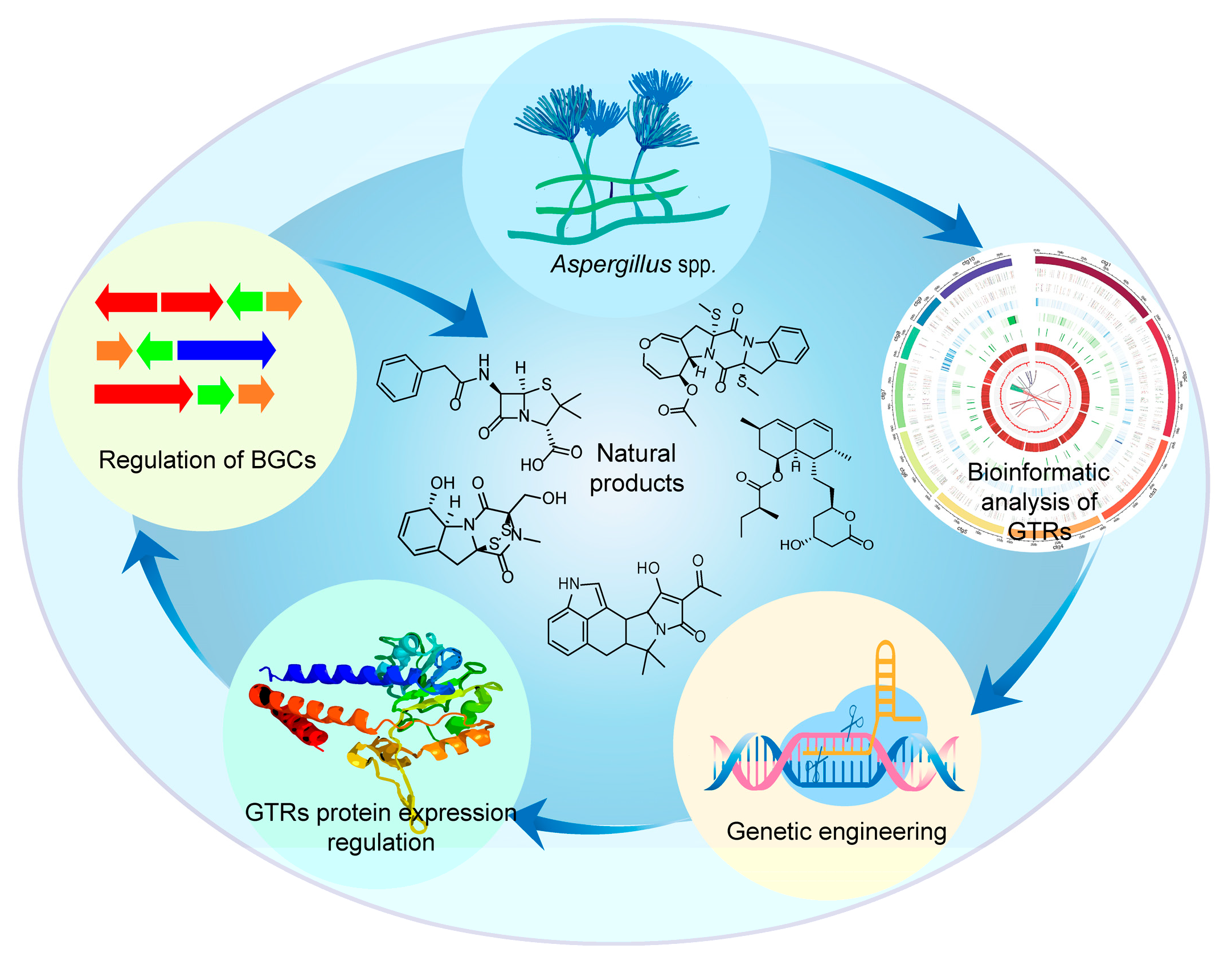
15. Xiaoze Bao*, Mengyuan Wu, Guishun Bai, Xusheng Duan, Jiangwei Pan, Yang Yang, Zeyu Wang, Xuanrong Sun, Jean Rodriguez, Damien Bonne*, Hong Wang*. Organocatalytic Diastereo- and Enantioselective Synthesis of Chlorinated and Trifluoromethylated Pyrrolidines Featuring a Removable Nitro Group. Organic Letters, 2025, 27: 7908-7913. (https://pubs.acs.org/doi/10.1021/acs.orglett.5c02233) IF: 5.0
Abstract
A new concept was reported to use nitro as a removable activating group for the stereoselective construction of chlorinated stereogenic centers. This was proven by the enantioselective (3 + 2) cycloaddition of isatin-derived 1,3-dipoles and chloronitroalkenes, affording a series of spiro-oxindoles featuring four consecutive stereogenic centers including one that is chlorinated.

16. Wei Hou, Xiaohui Zhou, Zhikun Yang, Huhui Xia, Yan Wang, Keren Xu, Shaoneng Hou, Shuning Zhang, Dongmei Cui, Peixiang Ma, Wei Zhou, Hongtao Xu. Multicomponent Reaction Integrating Selenium(II)–Nitrogen Exchange (SeNEx) Chemistry and Copper-Catalyzed Azide–Alkyne Cycloaddition (CuAAC). Angewandte Chemie International Edition, 2025, 64(16): e202500942. (https://doi.org/10.1002/anie.202500942) IF: 16.9
Graphical Abstract
1,4,5-Trisubstituted 5-seleno-1,2,3-triazoles were synthesized regioselectively with high atom economy by seamlessly integrating selenium(II)–nitrogen exchange (SeNEx) chemistry with CuAAC. This multicomponent reaction was applied to the successful identification of a potent Escherichia coli -glucuronidase inhibitor through target-based screening. A mononuclear σ-bound copper(I)-acetylide complex was synthesized and used to elucidate the reaction mechanism.

17. Weilong Cui, Luoqiang Zhang, Chu Liu, Junjia Su, Hao-Ran Zhang, Ronggang Ma, Daiqiang Jian, Wanxing Xiang, Xinyi Ye, and Xin Zhang. Reagent-Regulated Organocatalytic Enantiodivergent Synthesis of Chiral Sulfinimidate Esters. J. Am. Chem. Soc, 2025, 147(23): 19986–19995. (https://doi.org/10.1021/jacs.5c05035) IF: 15.6
Abstract
Achiral parameter-regulated enantiodivergent synthesis of chiral sulfur compounds has rarely been reported. Driven by the increasing importance of aza-sulfur stereogenic centers in drug discovery, we present herein organocatalytic tandem enantioselective chlorination and nucleophilic substitution for the synthesis of chiral sulfinimidate esters. The enantiopreference is modulated by N-chlorophthalimide (NCP) and trichloroisocyanuric acid (TCCA) with the same enantiomer of chiral pentanidium catalyst, and both enantiomers of sulfinimidate esters are obtained with high enantiocontrol. Notably, the reactions with TCCA are very fast and completed in 5 min, and the efficiency is well maintained for gram-scale synthesis. The resulting sulfinimidate esters are versatile precursors for diverse aza-S(IV) and S(VI) stereogenic centers. Studies on the enantiodivergent mechanism reveal that the sulfur stereogenic center is inverted twice in reactions with NCP, and only a single inversion is verified with TCCA, representing a novel strategy for enantiodivergent catalysis.

18. Wentao Wu,# Chu Liu,# Choon-Hong Tan,* and Xinyi Ye*. The Role of Anions in Guanidinium-Catalyzed Chiral Cation Ion Pair Catalysis. Acc. Chem. Res, 2025, 58: 2269−2281. (https://doi.org/10.1021/acs.accounts.5c00283) IF: 17.7
Abstract
Catalysts drive asymmetric transformations by orchestrating a network of covalent and noncovalent interactions that precisely regulate the reactivity and stereoselectivity. Ion pair catalysis, developed based on the inherent strength and long-range nature of ionic interactions, has demonstrated high catalytic efficiency and broad applicability. While chiral cationic catalysts have long been central to this field, the critical roles of their counteranions have historically been overlooked. Over the past 15 years, we have developed a class of N-sp2-hybridized guanidinium chiral cation ion pair catalysts, which have been widely applied in enantioselective reactions. In this Account, we present new insights into these catalysts, revealing how the roles of anions, acting as substrates, reagents, and cocatalysts, can be strategically leveraged to achieve remarkable enantioselectivity across a wide range of organic transformations.
When acting as substrates, anions, such as sulfinates, thiocarboxylates, and azides, are rendered reactive through intimate ion pairing with the chiral guanidinium moiety. This strategy facilitates desymmetrization processes, exemplified by the conversion of sulfinates to enantioenriched sulfinate esters and the remote desymmetrization of cis-dibromocyclohexanone via sequential SN2 and acyl transfer steps. Mechanistically, halogenophilic SN2X pathways (e.g., thiocarboxylate substitutions at sterically hindered tertiary carbons) bypass traditional steric limitations, while dynamic kinetic resolution of racemic bromides via azide substitution highlights the interplay between ion exchange and interfacial dynamics.
Anions generated in situ from stoichiometric reagents give rise to highly reactive intermediates such as enolates, sulfenates, and hypervalent silicates, which form ion pairs with chiral cations, enabling enantioselective transformations. For instance, enolates displace tertiary bromides via an SN2X mechanism (frontside attack), circumventing steric hindrance. Sulfur alkylation of sulfenamides yields chiral sulfilimines, while fluoride-activated acylsilanes undergo Brook-like rearrangements through penta-coordinate silicates. Silicon hydrides activated by fluoride form hydridosilicates, enabling enantioselective conjugate reductions of chromones and coumarins. The versatility of ion pairing is further illustrated by α-cyano carbanions in Pd-catalyzed decarboxylative allylic alkylations and is extended to a cooperative catalytic system, where DMAP-generated nucleophiles enable enantioselective phospha-Michael additions via dynamic cation-exchange activation.
The utilization of inorganic anions as cocatalysts further expands the scope of chiral cation ion pair catalysis. Peroxytungstate anions synergize with chiral cations to enable the epoxidation of allylic amines, while peroxomolybdate facilitates the N-oxidation of tertiary amines with high enantioselectivity. Beyond inorganic systems, organic anions also serve as effective cocatalysts. Notably, preformed pentanidium pyridinyl-sulfonamide ion pairs have been shown to catalyze the enantioselective Steglich rearrangement with high efficiency. This strategy extends to bisguanidinium sulfonated-phosphine/Pd ion pair catalysis, achieving stereocontrol in allylic amination of Morita-Baylis-Hillman substrates.
By systematically delineating the diverse roles of anions in guanidinium ion pair catalysis, this Account highlights new mechanistic insights and synthetic applications, paving the way for further advancements in asymmetric catalysis through precise control of ionic interactions.

19. Zhikun Yang #, Leyi Ying #, Ning Yang, Xue Yang, Hui Yan, Jiajia Han, Yanhong Yang, Xiaozhou Mou, Hong Wang*, Yasheng Li*. Rational discovery of novel tetrahydroisoquinoline-indole derivatives as potent P-glycoprotein inhibitor against multidrug resistance in MCF-7/ADR cell. Bioorganic Chemistry, 2025, 164: 108854. (https://doi.org/10.1016/j.bioorg.2025.108854) IF: 4.7
Abstract
A key strategy for overcoming multidrug resistance (MDR) involves developing inhibitors for the chemotherapy drug efflux pump, P-glycoprotein (P-gp). In this study, building on the prior exploration of the tetrahydroisoquinoline-indole scaffold and receptor-based drug design, the amino group on the indole underwent nucleophilic substitution, leading to 20 novel tetrahydroisoquinoline-benzyl-1H-indole derivatives. The exploration of structure-activity relationships (SAR) revealed that compound BP3p exhibited low cytotoxicity and potent MDR reversal activity against doxorubicin in MCF7/ADR cells (IC50 = 0.11 μM, reversal fold = 215.9), surpassing the reported P-gp inhibitor cyclosporine A. Furthermore, mechanistic studies indicate that BP3p's MDR reversal activity stems from inhibiting the efflux function of P-gp but not expression, and BP3p could bind to P-gp directly and induced a conformation change of P-gp. Moreover, molecular docking simulations further confirm that BP3p shows a strong affinity for P-gp, while 3D tumor spheroid models suggest that BP3p exhibits significant reversal activity against doxorubicin-resistant cells, underscoring its potential to overcome tumor resistance in vivo. These findings position BP3p as a promising P-gp inhibitor, offering valuable insights for the future development of novel P-gp inhibitors.
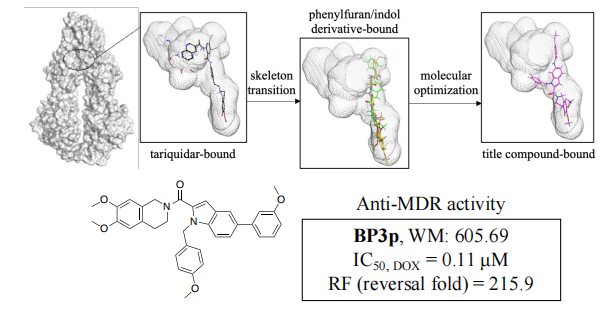
20. Yunpeng Wang, Zilin Gao, Kun Fang, Yongqiang Tu*, Sihua Hou, Hong Wang, Ka Lu, Baoheng Dou, Jun Xue, Tian Ke, Shuyu Zhang, Zhimin Chen, Tongmei Ding. Construction of Fused–Bridged Tricycles by Molecular Reorganization Involving Prins Cyclization/Dicycloexpansion. Angew. Chem. Int. Ed. 2025, e202514340. (https://doi.org/10.1002/anie.202514340)IF: 16.90
Abstract
The development of efficient methods for constructing structurally complex and diverse polycyclic carbon frameworks remains in high demand for the synthesis of pharmaceuticals and bioactive natural products. One such method lies in engineered one-step molecular reorganization through the cascade cyclization/cycloexpansion of rationally designed precursors with interconnected small-ring systems. Here, we have strategically designed and developed a synthetically valuable method to construct fused-bridged tricyclic systems through cascade Prins cyclization/dicycloexpansion of substrates with a cycloalkylidene (CAD) tethered to a cycloketones. This methodology features high step economy, reaction efficiency, and selectivity, as well as a broad substrate tolerance. Diversification through variation of the size of the small ring, the linker length, and the linking site has enabled the efficient synthesis of six types of fused-bridged tricyclic scaffolds, which are found frequently in bioactive natural products. Based on supporting experiments and DFT calculations, a plausible reaction mechanism is also proposed.

21. Zhaochao Wang, Juanjuan Yu, Chenjie Wang, Yi Hua, Hong Wang*, Jianwei Chen*. The deep mining era: Genomic, metabolic, and integrative approaches to microbial natural products from 2018 to 2024. Marine Drugs, 2025, 23, 261. (https://doi.org/10.3390/md23070261)
22. Gang-Ao Hu#, Huai-Ying Sun#, Qun-Jian Yin, He Wang, Shi-Yi Liu, Bin-Gui Wang, Hong Wang, Xin Li*, Bin Wei*. Exploring the biosynthetic potential of microorganisms from the South China Sea cold seep using culture-dependent and culture-independent approaches. Marine Drugs (2025), 23: 313.
Abstract
Cold seep ecosystems harbor unique microbial communities with potential for producing secondary metabolites. However, the metabolic potential of cold seep microorganisms in the South China Sea remains under-recognized. This study employed both culture-dependent and culture-independent approaches, including 16S rRNA amplicon sequencing and metagenomics, to investigate microbial communities and their potential for secondary metabolite production in the South China Sea cold seep. The results indicate microbial composition varied little between two non-reductive sediments but differed significantly from the reductive sediment, primarily due to Planctomycetes and Actinobacteria. Predicting the Secondary Metabolism Potential using Amplicon (PSMPA) predictions revealed 115 strains encoding more than 10 biosynthetic gene clusters (BGCs), with lower BGC abundance in reductive sediment. Culture-dependent studies showed Firmicutes as the dominant cultivable phylum, with strains from shallow samples encoding fewer BGCs. Metagenomic data confirmed distinct microbial compositions and BGC distributions across sediment types, with cold seep type having a stronger influence than geographic location. Certain BGCs showed strong correlations with sediment depth, reflecting microbial adaptation to nutrient-limited environments. This study provides a comprehensive analysis of the metabolic capabilities of South China Sea cold seep microorganisms and reveals key factors influencing their secondary metabolic potential, offering valuable insights for the efficient exploration of cold seep biological resources.

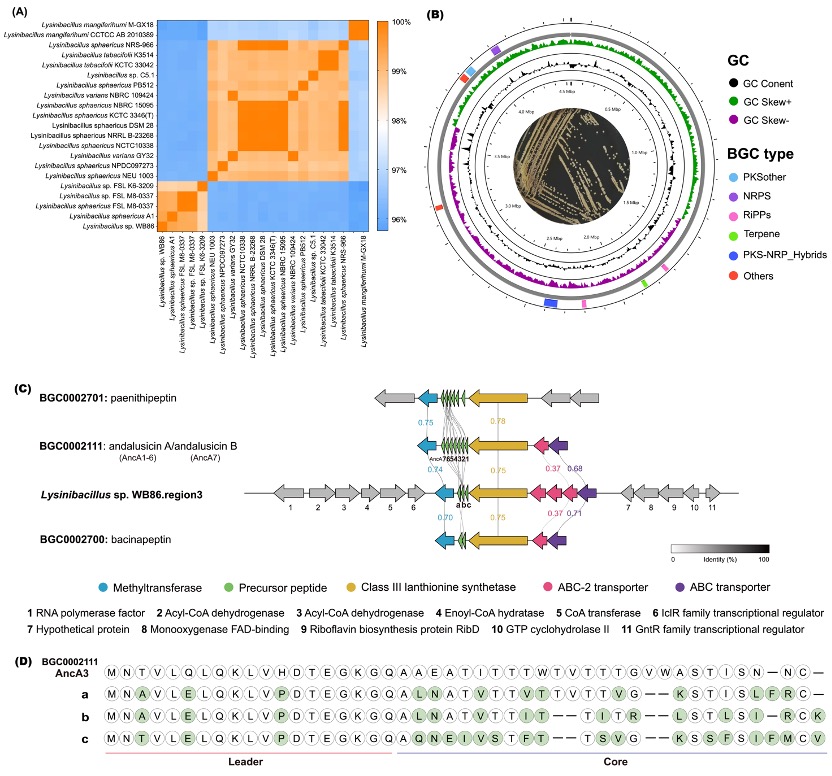
23. Huai-Ying Sun, Ji-Xiang Xin, Gen Li, Zhen-Yi Zhou, Xin Li, Hong Wang, Qun-Jian Yin*, Bin Wei*. Complete genome sequence of Lysinibacillus sp. WB86, a potential antimicrobial lanthipeptides producer. Marine Genomics (2025), 82: 101204.
Abstract
Lysinibacillus sp. WB86 was isolated from a cold seep in the South China Sea, and its complete genome was sequenced using Oxford Nanopore Technologies (ONT). The genome consists of a single circular chromosome spanning 4,537,071 bp, with a G+C content of 37%. FastANI analysis revealed a 99.38% average nucleotide identity (ANI) with Lysinibacillus sphaericus A1, confirming its classification as a strain of L. sphaericus. Genome annotation identified 4,397 protein-coding genes, along with 111 tRNAs and 34 rRNAs. Functional categorization using the COG database assigned 84.6% of the protein-coding genes to specific roles, with the majority involved in transcription, amino acid transport and metabolism, translation and ribosomal biogenesis, and replication, recombination, and repair. Additionally, antiSMASH-based analysis uncovered eight biosynthetic gene clusters (BGCs), only one of which exhibited high similarity (>50%) to known BGCs. This cluster was predicted to encode novel antimicrobial lanthipeptides, suggesting potential biotechnological applications.
24. Qing-Hong Meng, Yu-Tian Guan, Jing-Hai Wang, Xing-Yan Wang, Xiang-Mei Ren, Hai-Yan Sui, Bin Wei*, Xiao-Ying Bian*. Genome mining of Chromobacterium genus reveals a class of nonribosomal lipopeptides with potent antifungal activity. Journal of Agricultural and Food Chemistry (2025). doi: 10.1021/acs.jafc.5c04892.
Abstract
Bacterial genomes encode numerous cryptic biosynthetic gene clusters (BGCs) that represent an untapped potential source of drugs or biopesticides. However, the limited analysis of BGCs in a specific genus has limited the discovery of their natural products through genome mining. Herein, we report the systematic analysis of the biosynthetic potential of 136 genomes of the Chromobacterium genus. A total of 1713 BGCs were identified and grouped into 190 gene cluster families (GCFs). However, only eight BGCs from seven GCFs have been functionally characterized, highlighting the vast underexplored potential of the genus for the production of novel natural products. Guided by this analysis, we investigated a cryptic BGC of GCF9 featuring a glycosyltransferase and a nonribosomal peptide synthetase (NRPS) with a starter condensation domain, leading to the identification of a class of glycolipopeptides chromorhipeptins from Chromobacterium rhizoryzae. Chromorhipeptins A and B demonstrated broad-spectrum antifungal activity against several plant pathogenic fungi, including Valsa mali, with minimum inhibitory concentrations (MICs) at 0.04 and 0.16 μM, respectively, outperforming the fungicide carbendazim (MIC = 0.78 μM). These findings revealed the biosynthetic potential of Chromobacterium and underscored the power of genome mining to unlock cryptic bacterial natural products for crop protection against plant pathogenic fungi.

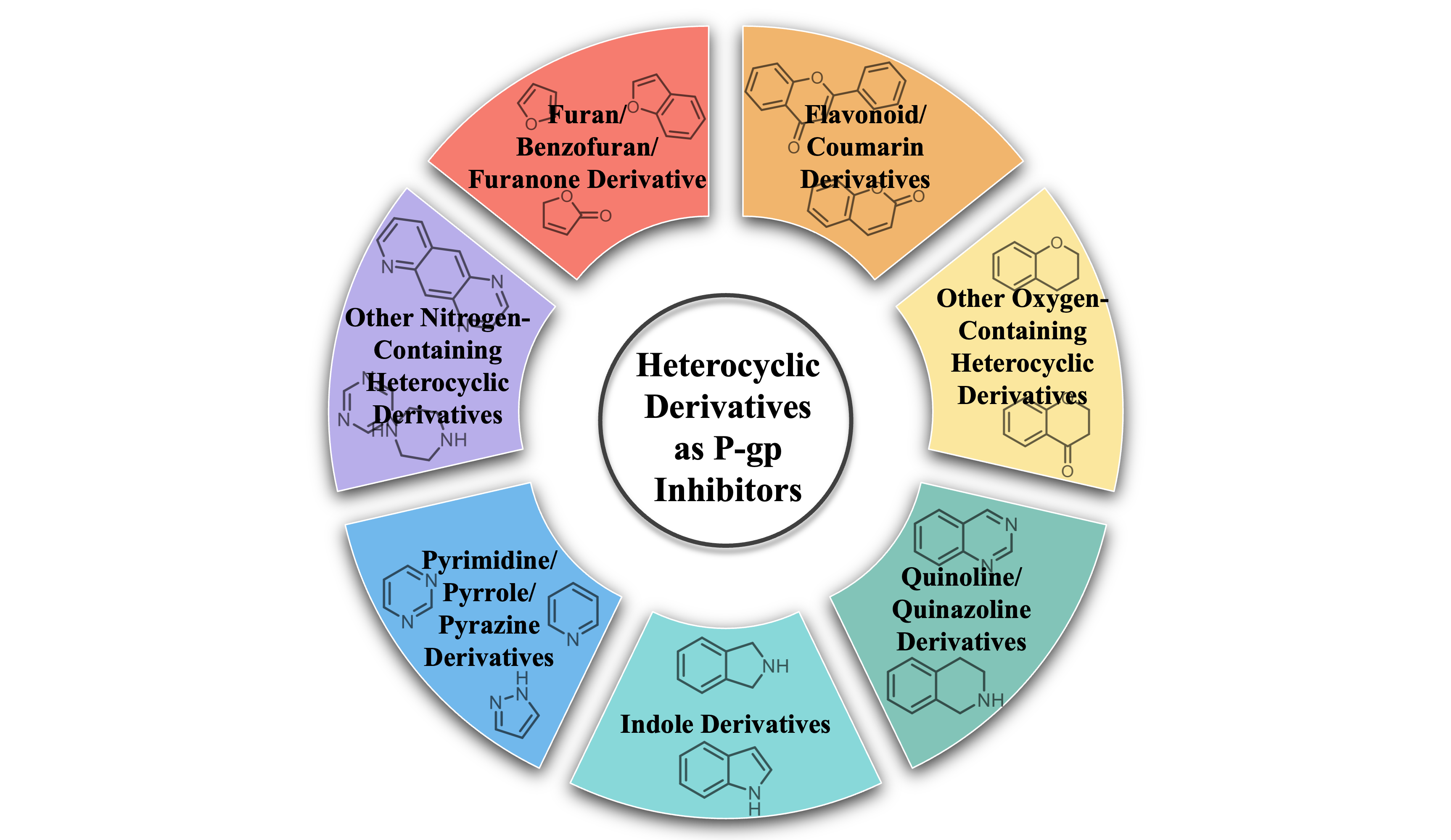
25. Zhikun Yang, Yanhong Yang, Zimeng Huang, Yi Hua, Mahmoud Emam Abd El-Salam Hassaan, Hong Wang. Development of heterocyclic derivatives as P-glycoprotein inhibitors against multidrug resistance: pharmacological activities, structure–activity relationship and target (2020–2024). RSC Medicinal Chemistry (2025). doi: 10.1039/d5md00609k.
Abstract
The overexpression of P-glycoprotein (P-gp) has been recognized as a pivotal factor contributing to the emergence of multidrug resistance (MDR), a phenomenon that frequently limits the efficacy of chemotherapy and profoundly impacts patient prognosis. Consequently, the inhibition of P-gp's efflux function has become a critical therapeutic strategy for overcoming drug resistance and enhancing chemotherapeutic efficacy. In recent years, the development of P-gp inhibitors has garnered significant attention, particularly with the frequent incorporation of heterocyclic derivatives, which exhibit exceptional biological activity and favorable chemical properties, into drug design. In this paper, we reviewed the latest research progress of pharmacological activities, structure-activity relationships and molecular targets of heterocyclic derivatives as P-gp inhibitors in the past five years (2020-2024). Through this comprehensive analysis, the potential of heterocyclic derivatives in modulating P-gp inhibition is highlighted, positioning them as promising candidates for the development of novel anti-resistance therapeutics. Meanwhile, the review offers a solid theoretical foundation and experimental guidance for the future design of more efficacious P-gp inhibitors.
26. Yanlei Yu, Yunjie Zhao, Honggang Wang, Yi Hua, Wenchao Yu, Ningning Wang, Bin Wei, Hong Wang*. A low molecular weight fucoidan with high sulfation has anti-obesity activity via gut microbiota regulation. Carbohydrate Polymers. 2025, 364, 123739.
Abstract
Obesity is a chronic disease that can lead to various health complications, including diabetes, hypertension, and cardiovascular disease. Fucoidan has been identified as an effective agent in prevention and mitigation of obesity. However, its absorption and bioavailability are significantly affected by its high molecular weight and poor solubility. In this study, we present a low molecular weight fucoidan, designated as LMWF4, derived from Saccharina japonica, which has a molecular weight of 2.6 kDa and a high sulfation content of 37.7 %. The structural backbone of LMWF4 consists of α-(1 → 3)-L-fucose, with sulfation modifications primarily occurring at the C2 or C4 position. Mouse experiments have demonstrated that LMWF4 exhibits anti-obesity activity by mitigating the reduction of the α-diversity index of intestinal flora induced by a high-fat diet. Furthermore, LMWF4 significantly increased the relative abundance of microbial taxa negatively associated with obesity, including g_Akkermansia;s__uncultured_bacterium and g__Blautia;s__Lachnospiraceae_bacterium. Conversely, LMWF4 was found to decrease the relative abundance of g__Rikenellaceae_RC9_gut_group, which is positively correlated with obesity. Our discovery indicates that modifications characterized by relatively low Mw and a high sulfation degree exhibit a more pronounced anti-obesity effect. Additionally, these modifications demonstrate a greater capacity to regulate intestinal flora disorders, thereby presenting a potential candidate for anti-obesity treatment.
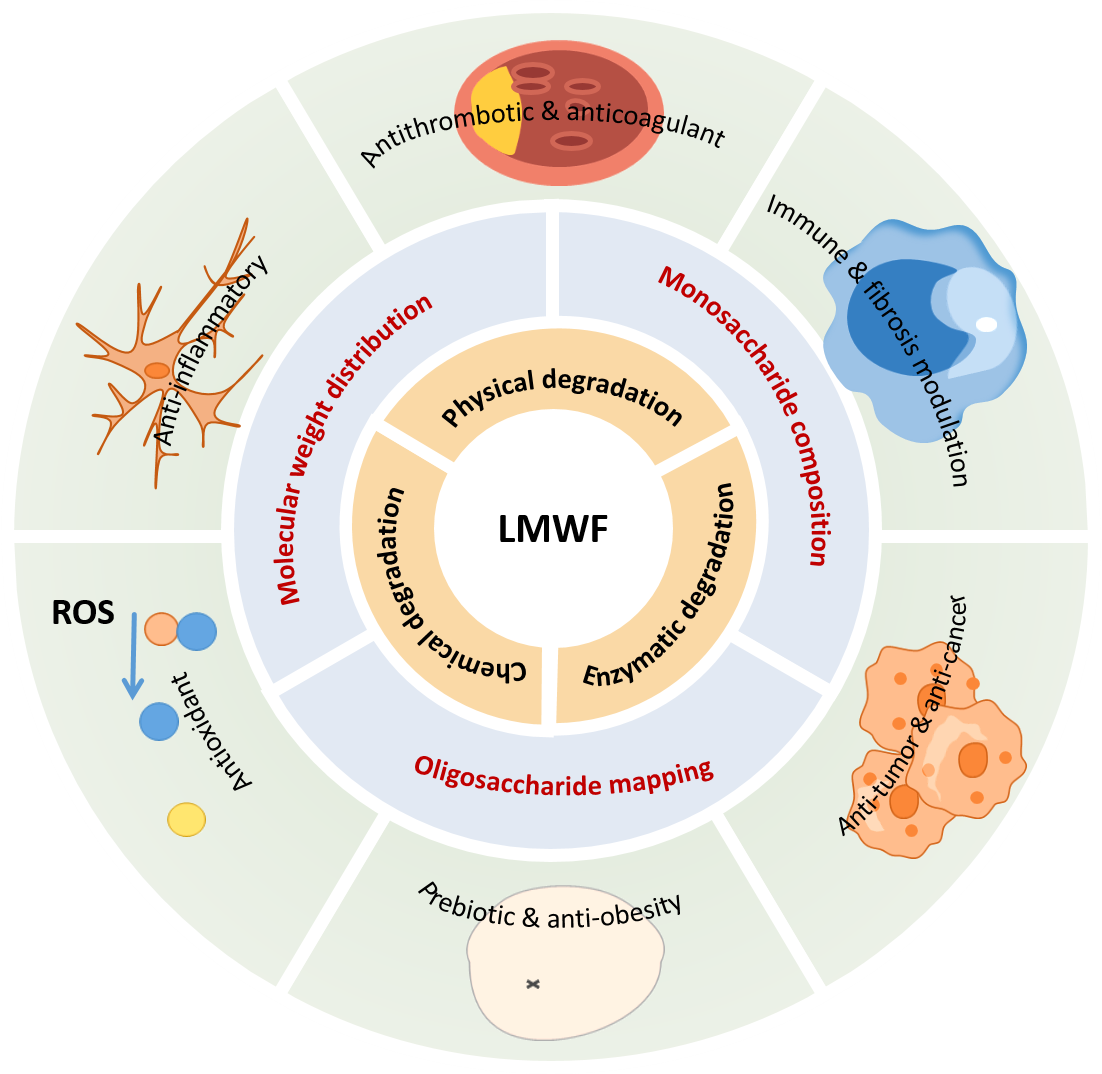
27. Yunqi Yang, Ningning Wang, Yi Hua, Bin Wei, Hong Wang, Yanlei Yu*. Developments in the analytical methodologies and structure-activity relationships of low molecular weight fucoidans: a review. International Journal of Biological Macromolecules. 2025, 328, 147544.
Abstract
Fucoidan is an acidic polysaccharide associated with a variety of biological functions, including anticoagulant, antithrombotic, immunomodulatory, and antitumor activities. It has been extensively utilized in both the pharmaceutical and food industries. However, the high intra- and intermolecular diversity of fucoidan poses challenges in correlating specific structures with biological functions and in developing practical applications. Low molecular weight fucoidan (LMWF), derived from fucoidan, reduces this complexity, facilitating structural refinement, characterization, and the assignment of structure to function. Additionally, LMWF offers several advantages, such as enhanced solubility, reduced viscosity, and improved absorption, which lead to increased bioavailability and promising potential for application. Nonetheless, a critical gap persists in the literature regarding the ongoing challenges associated with the structural elucidation of LMWF and its impact on bioactivity. Therefore, this review highlights recent advancements in structural characterization methodologies, with a particular emphasis on oligosaccharide mapping and the structure-activity relationship (SAR) of LMWF in relation to its biological functions. Additionally, it presents various techniques for preparing LMWF, including degradation and synthesis techniques. This review aims to serve as a comprehensive and timely reference for the rational design and application of LMWF-based biofunctional systems.
28. Wenchao Yu, Shuqi Ge, Jialian Huang, Bin Wei, Qihao Wu, Hong Wang*, Yanlei Yu*. Paired genomic and metabolomic analysis reveals the secondary metabolome potential of Cystobacter. Journal of Applied Microbiology. 2025 https://doi.org/10.1093/jambio/lxaf238
Abstract
Aims
The genus Cystobacterhas been reported to produce bioactive secondary metabolites and harborabundant biosynthetic gene clusters (BGCs). However its genomic and metabolomicpotential remains largely unexplored due to the lack of systematic data miningstudies. Here, we present the first paired genomic and metabolomic analysis of Cystobacterto uncover its secondary metabolic capabilities.
Methodsand results
By examining its genomic features and classifying BGCpatterns, we provided a comprehensive overview of the biosynthetic capabilitiesof Cystobacter. Our analysis revealed that 91% of BGCs remainuncharacterized, with ribosomally synthesized and post-translationally modifiedpeptides (RiPPs) being the most predominant class. An in-depth analysis of RiPP core peptides from Cystobacter demonstrateddistinct features compared to those from other species, and deep learningmodels predicted that five of these peptides have antimicrobial potential.As a proof of concept, we integrated the genomic data of Cystobacter ferrugineusCbfe23 with its metabolomics profile, leading to the correlation of acluster of novel tubulysin analogs with its biosynthetic pathway.
Conclusion
Thispaired genomic and metabolomic analysis reveals the untapped secondarymetabolic potential of Cystobacter, highlighting its important roles indrug discovery, microbial communities, and bioengineering.
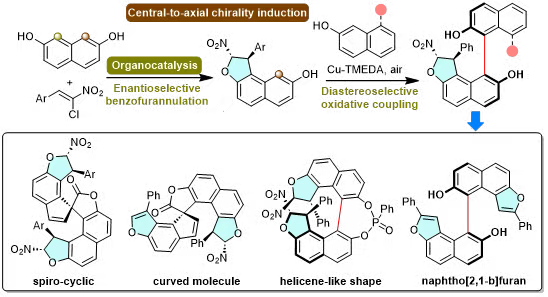
29. Yang Yang#, Guishun Bai#, Mengmeng Qin#, Juelian Wang, Yihuan Yang, Hong Wang, Damien Bonne, Jean Rodriguez, Xiaoze Bao*. Atroposelective synthesis of sterically hindered stereochemically complex BINOL derivatives via central-to-axial chirality induction. Chemical Science. 2025. (https://doi.org/10.1039/D5SC05212B)
Abstract
A sequential strategy is proposed for the atroposelective construction of new families of sterically hindered BINOL derivatives bearing multiple stereogenic elements and featuring up to four stereogenic centers. The sequence begins with an organocatalyzed regio- and enantioselective mono-dihydrobenzofurannulation of commercially available 2,7-dihydroxynaphthalene establishing two stereogenic carbon atoms, followed by highly atroposelective copper-catalyzed aerobic oxidative homo- or cross-couplings fixing the axial chirality thanks to a nearly perfect induction of chirality. In addition, the resulting BINOL derivatives serve as promising precursors for the synthesis of either complex spiroheterocycles by axial-to-central conversion of chirality, heterohelicene-like molecules by axial-to-helical conversion of chirality, or an atropisomeric naphtho[2,1-b]furan scaffold via 1,2-aryl migration, further underscoring the potential of this new atroposelective strategy based on a practical central-to-axial chirality induction.
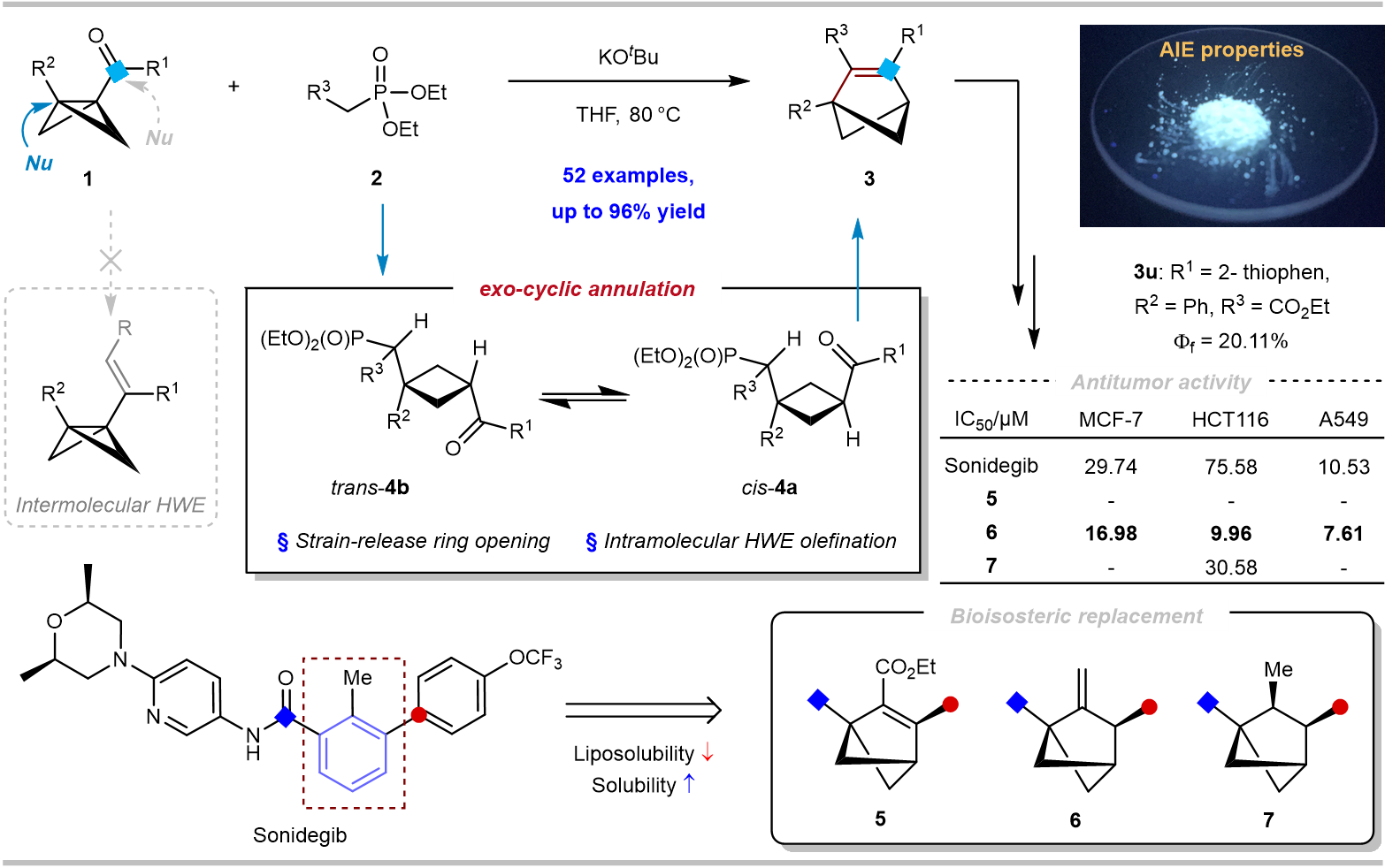
30. Junjie Ge#, Lihang Cao#, Kun Lin, Chenhao Ruan, Xinyi Ye, Xiaoze Bao, Zhikun Yang,* Hong Wang,** and Hua Chen***, Ring Expansion of Bicyclo[1.1.0]butyl Ketones to Bicyclo[2.1.1]hexenes Using Ketone as Both Activating and Reacting Groups. Nat. Commun. 2025, 16, 9190. (https://doi.org/10.1038/s41467-025-64226-z) IF: 15.7.
Abstract
Bicyclo[n.1.1]alkyl skeletons are of significant interest as bioisosteres of phenyl groups for lead drug modification. Common strategies for their synthesis utilize bicyclo[1.1.0]butanes, wherein the electron-withdrawing groups serve solely as activating groups to facilitate the cleavage of the bridged σ bond, leading to ring expansion by an insertion manner. In this work, we utilize ketones in bicyclo[1.1.0]butane as both activating and reacting groups, promoting a tandem nucleophilic addition/intramolecular Horner-WadsworthEmmons olefination process by a distinct exo-cyclic annulation manner. The approach enables the synthesis of diverse bicyclo[2.1.1]hexenes (52 examples, up to 96% yield). A series of transformations have been carried out on bicyclo[2.1.1]hexenes to synthesize bicyclo[2.1.1]hexane/hexene analogues of sonidegib, and the physicochemical properties and anti-tumor activities of these analogues have been investigated. In addition, this approach enables the discovery of aggregation-induced emission properties in this distinctive ring skeleton.
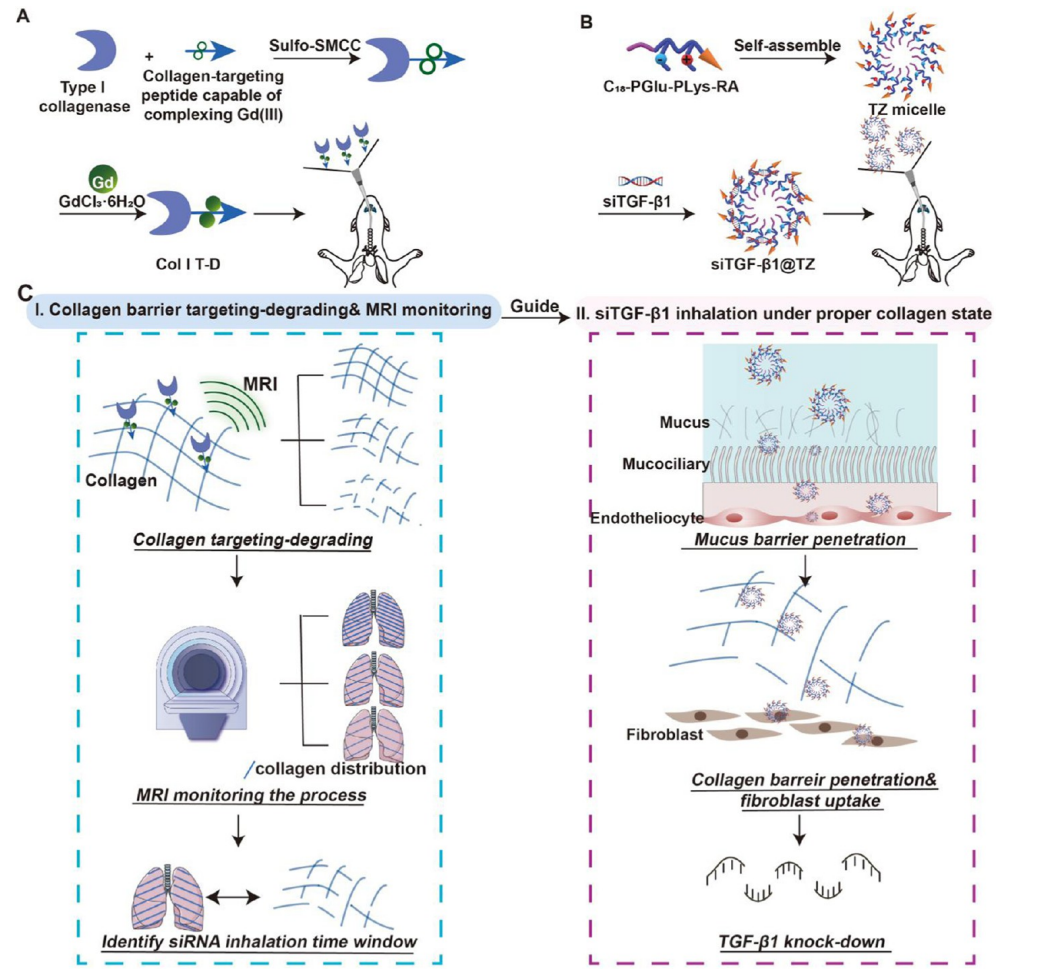
31. Jie Shen, Xinrui Zhang, Jie Wang, Xusheng Duan, junhao pan, Yue Cai, Bin Wei, Hong Wang, Xuanrong Sun*, Targeted collagen degradation by an MRI probe facilitates siRNA delivery for sequential theranostics in Pulmonary Fibrosis. ACS Nano. 2025, 19(14):14028-14043. (https://doi.org/10.1021/acsnano.4c18383) IF: 16.1
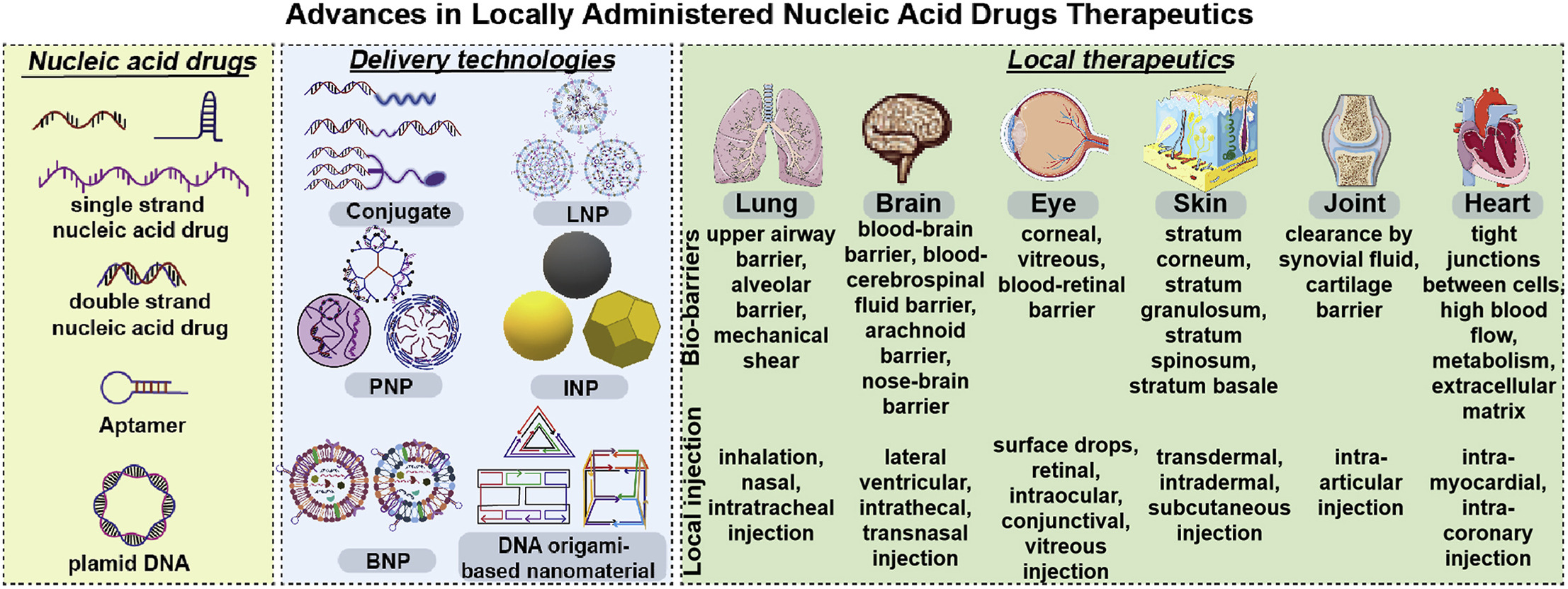
32. Jie Shen, Xusheng Duan, Ting Xie, Xinrui Zhang, Yue Cai, Junhao Pan, Xin Zhang, Xuanrong Sun*. Advances in Locally Administered Nucleic Acid Therapeutics. Bioactive Materials. 2025, 49:218-254. (https://doi.org/10.1016/j.bioactmat.2025.02.043) IF: 20.3
33. Yue Cai, Meilin Qiao, Xiuzhi Xie, Yujie Zhou, Ruibing Yu, Fuhang Song, Hong Wang, Huawei Zhang, Xuanrong Sun*, Intracellular MRSA-targeted lipid-polymer hybrid nanoparticles for macrophage reprogramming and intracellular MRSA eradication. Chemical Engineering Journal. 2025, 505:159665. (https://doi.org/10.1016/j.cej.2025.159665) IF:13.2
34. Chenjie wang, Cancan Wang, Si Chen, Shi Kai, Juanjuan Yu, Yiping Ding,Yujie Yue, Yi Hua, Hong Wang, Jianwei Chen*. Global landscape of antibiotic resistance genes in the human gut microbiome metagenome-assembled genomes. BMC Microbiol. 2025. (https://doi.org/10.1186/s12866-025-04586-0)